Geneviève Arsenault-Labrecque, Postdoctoral Fellow, Vanessa Tremblay, Research Professional and Richard Bélanger, Professor and Research Chair in Plant Protection, Université Laval – Spring Pulse Beat 2022

SINCE THE MID-1950S, many soybean growers across Canada have struggled with Phytophthora root rot (PRR), caused by the oomycete Phytophthora sojae. Once the disease is present in their fields, farmers must deal with it for a long time as surviving spores will persist in the soil for approximately 10 years.
Unlike other diseases that are easier to recognize in the field, such as white mould (Sclerotinia stem rot), PRR is an insidious soil- borne pathogen that can affect plants at every growth stage. Where the disease is established, it can cause an average of $160 per hectare in yield losses every season while often going undetected. Symptoms of PRR range from pre- and post-emergent damping off, stem and root rot, stunted plant growth, wilted plants and even plant death. In North America, annual losses attributed to this disease can exceed $500 million.
Different methods are used to fight off PRR. Seed treatments will be efficient in the early season to prevent damping-off, but protection will be lost as soon as the roots emerge. Tolerant varieties can also be used to diminish the impact of the disease, but it is well established that Rps (resistance to Phytophthora sojae) genes will confer complete protection of plants from seedling to harvest when matched against specific isolates of P. sojae.
Indeed, when soybean varieties carrying gene Rps1a were first deployed in Canada almost 60 years ago, the disease was under control until the emergence of P. sojae isolates with a pathotype able to circumvent Rps1a. Rps1c was then deployed in 1979 and has been effective for several years. Unfortunately, its intensive use in Canadian soybean fields ultimately led to the emergence of new pathotypes of P. sojae and a declining efficiency of this source of resistance. This has led to the deployment of new Rps genes so that nowadays, five different Rps genes can be found in commercial soybean lines: Rps1a, Rps1c, Rps1k, Rps3a and Rps6. With the durability of Rps genes in the field estimated to last 8–20 years, breeders also have new resistance genes in their sights, including Rps8 and Rps11.
With the deployment of these various sources of resistance, P. sojae has developed new variants (pathotypes) and more than 200 can be found worldwide. Without the possibility of knowing which pathotypes are found in their fields, it has become challenging for growers and seed companies to know what Rps genes will be efficient to curb PRR. Phenotyping assays can be used to assess P. sojae pathotypes, but on a large scale, it is a time-consuming, labour-intensive and often imprecise method. For this reason, a molecular tool using precise genetic markers was developed in our lab that allows for rapid identification of P. sojae pathotypes found in a field. This allows farmers to choose Rps genes that will be efficient for the next growing season.
The efficiency of this new molecular assay was recently demonstrated in a pan-Canadian survey that also aimed to characterize the diversity of P. sojae pathotypes in Canada. For this survey, soil samples from a total of 246 fields were collected during the growing seasons of 2016 to 2019, including 84 in Manitoba, 81 in Ontario and 81 in Québec. A total of 295 unique isolates of P. sojae were obtained, 107 originating from Manitoba, 96 from Ontario and 92 from Québec. A subsample of those isolates was first phenotyped with our traditional hydroponic assay and genotyped in parallel with the newly developed molecular assay to validate it. Once the molecular tool efficiency was confirmed (98.6%), it was used to characterize the remaining isolates. Upon analysis, 24 different pathotypes were detected in Québec and Ontario, and eight were detected in Manitoba, where soybean production is more recent.
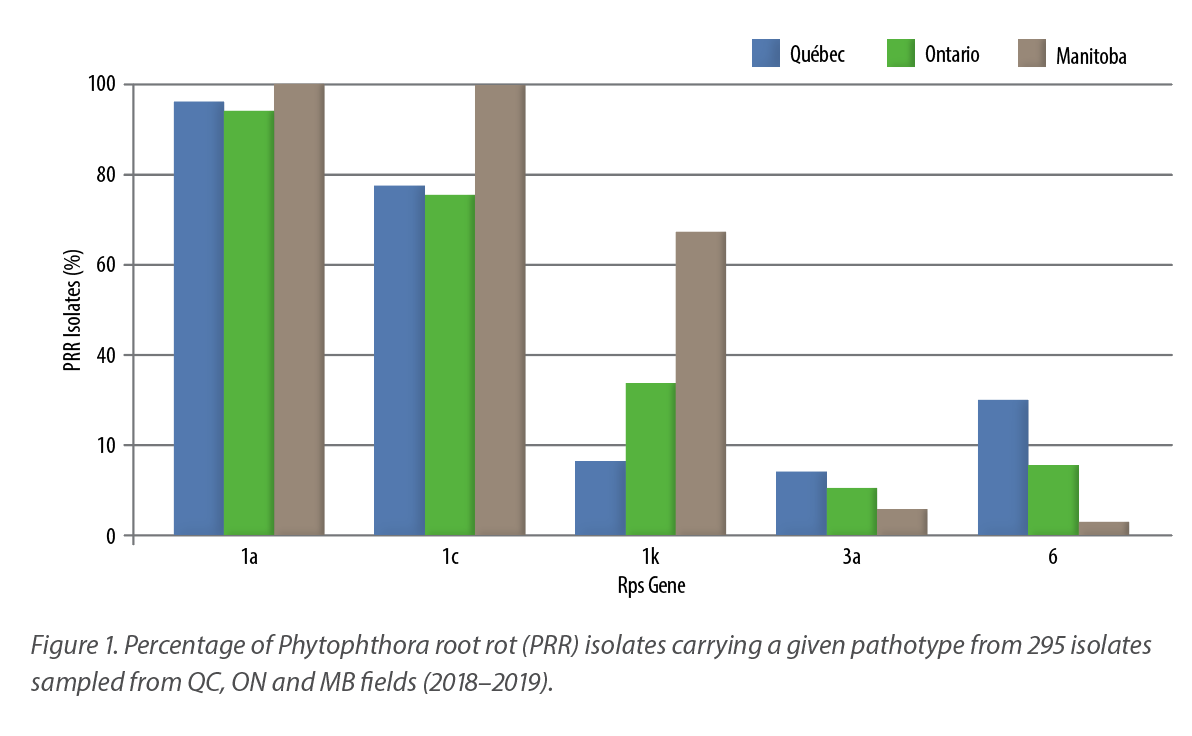
As shown in Figure 1, this study also highlighted that some sources of resistance to PRR in soybeans are rapidly declining. For instance, Rps1a was overcome by 96, 94 and 100% of P. sojae isolates tested from Québec, Ontario and Manitoba, respectively. Rps1c followed closely, as it was overcome by 77, 75 and 100% of the isolates, respectively. Concerning Rps1k, its efficiency is somehow maintained in Québec and Ontario, with 16 and 33% of the isolates able to circumvent it, respectively, while this percentage rose to 67% in isolates from Manitoba. For Rps3a and Rps6, less than 20% of the isolates are able to circumvent it across Canada, with the exception of Rps6 in Québec, where almost one isolate out of three is able to overcome its resistance.
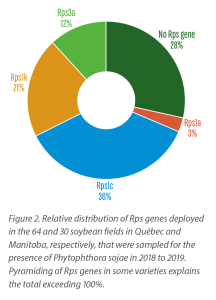
During the course of this survey, soybean varieties that were grown in the sampled fields from Manitoba and Québec were recorded. As shown in Figure 2, Rps1c was the most common resistance gene used, followed by Rps1k, Rps1a and Rps3a, while nearly one-third of the fields were planted with cultivars carrying no Rps genes. When data was compared with the P. sojae pathotypes detected in a given field, it was found that 84% of farmers grew soybean varieties that were not resistant against the pathotypes of the isolates found in the fields.
These results stress the importance of carefully managing the declining sources of resistance in light of the widespread use of resistance genes, Rps1a and Rps1c, since their deployment in the 1950s. In order to do that, growers should select resistant soybean varieties according to the pathotypes of the P. sojae isolates that are found in their field to ensure their efficiency and preserve the longevity of these resistance sources. It is also essential for breeders to direct their efforts to Rps genes that are still efficient (Rps1k, Rps3a and Rps6) while developing resistant varieties, without losing sight of promising genes such as Rps8 and Rps11.
Growers already have the opportunity to detect PRR presence from soil or plant samples and to characterize P. sojae pathotypes in order to know which Rps genes they should use. This service is offered by AYOS diagnostic, a spinoff founded by students in the lab that aims to ensure the developed diagnostic tool is available for all Canadian growers. They are also working on a similar tool with AAFC that targets soybean cyst nematode and its two current sources of resistance: Peking and PI 88788. ■